Spatial and Spatiotemporal Point Process Modelling in Epidemiology
by IDReC

TILMAN M. DAVIES
PhD candidate, Dept. of Statistics.
Epidemiologists often deal with data sets comprised of point locations of the disease(s) being studied in order to gain an insight into the spatial variation and/or correlation of infections over a given geographical region. A critical component of this process is the availability of sound statistical methods able to cope with the complex trends and dependence structures typically present in such datasets. My research has dealt with the appraisal and refinement of certain point process methodologies with a view to improved modelling of epidemiologically-flavoured problems.
Kernel-Smoothed Density-Ratios
An expression of the ‘risk' of infection is a useful statistic commonly employed by epidemiologists. Spatially, this can be achieved by nonparametrically estimating the 2-dimensional probability density functions related to the recorded ‘case' and ‘control' location data via kernel smoothing, and evaluating their ratio. We improved the method by considering variable smoothing in the estimates, and developing computationally efficient methods of highlighting statistically significant sub-regions of risk.
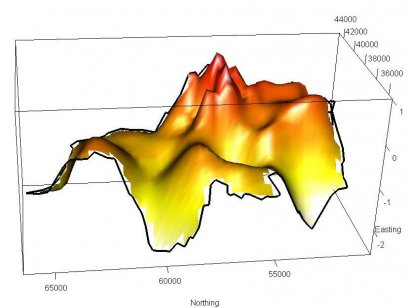
Figure 1. A surface plot showing the relative risk of liver disease in North East England.
Spatial Point Processes Models
A more detailed investigation into the behaviour of the observed point patterns can be achieved if we consider the presence of inter-point correlation. We investigated the particularly powerful class of models known as ‘log-Gaussian Cox processes,' (LGCP) capable of tracking the evolution of the spatiotemporal disease intensities. This work highlighted the potential the LGCP has in epidemiological applications, and established novel numerical results related to the fitting and conditional simulation of these models. We also look at shot noise process models, which allow simulation of putative centres of heightened risk.
Animation 1. Video of Metropolis-Hastings birth -death algorithm performing spatial simulation of possible locations of centres of disease risk. Note that disease need not always develop close to such centres.
Computer software
The lack of accessibility to some of the more complicated statistical methods can often be a problem in terms of encouraging their use. Alongside the theoretical and empirical work completed with respect to the kernel density-ratio and LGCP, we developed and released freely available software (LGCP package developed in collaboration with leading researchers at Lancaster University, UK) for use with the statistical computing environment ‘R' to perform operations related to the above topics. The availability of these software packages has already yielded interesting collaborative work with epidemiologists and other scientists around the globe.
The R packages that Tilman has helped to develop are available on the links below:
Spatial relative risk - sparr
log-Gaussian Cox process - lgcp
Tilman has recently been examined on his doctoral thesis and has taken a lectureship post within the Maths and Statistics Department at the University of Otago. Congratulations Tilman, and we wish you the best for this next stage of your career.